# synapsis ## Project overview synapsis is an R package for analysing fluorescent microscopy images. ## Contributors Lucy McNeill, St Vincent's Institute of Medical Research Wayne Crismani, St Vincent's Institute of Medical Research and the University of Melbourne ## Compatibility ## Using synapsis synapsis has four main functions. These are: - auto_crop - get_pachytene - count_foci - measure_distances We summarise them in the following subsections: ### auto_crop input: Original grey scale image files of (1) Synaptonemal complexes (e.g. SYCP3 anti-body) and (2) Foci (e.g. MLH1, MLH3 anti-body) channels from e.g. Nikon .nd2 files. output: crops in channels (1) '*dna.jpeg' and (2) '*foci.jpeg' around individual cells. 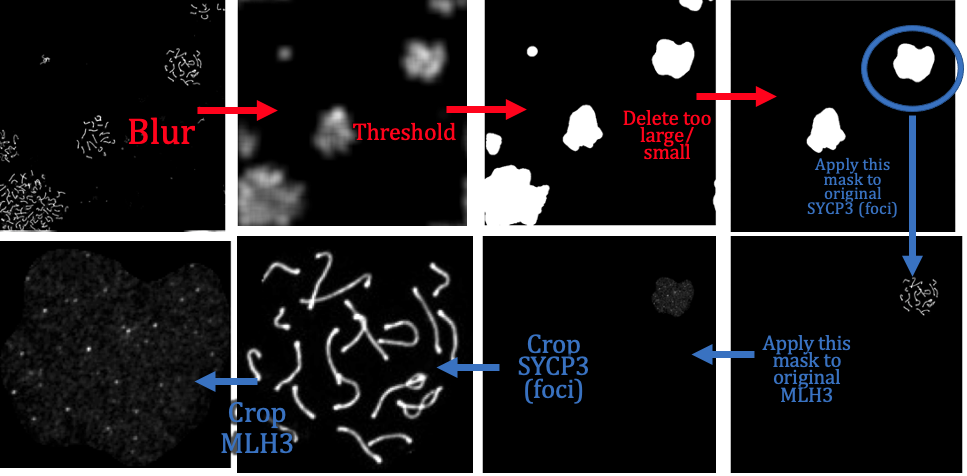 ### get_pachytene input: crops in channels (1) '*dna.jpeg' and (2) '*foci.jpeg' around individual cells, from previous auto_crop. output: only keeps crops if cells are in pachytene phase (based on channel (1)) ### count_foci input: crops of dna and foci channels in pachytene phase (from get_pachytene) output: number of foci counts of synamtonemal complexes per cell (i.e. channel 1 coincident with channel 2) as a function of genotype. ### measure_distances input: output: ## Analysis 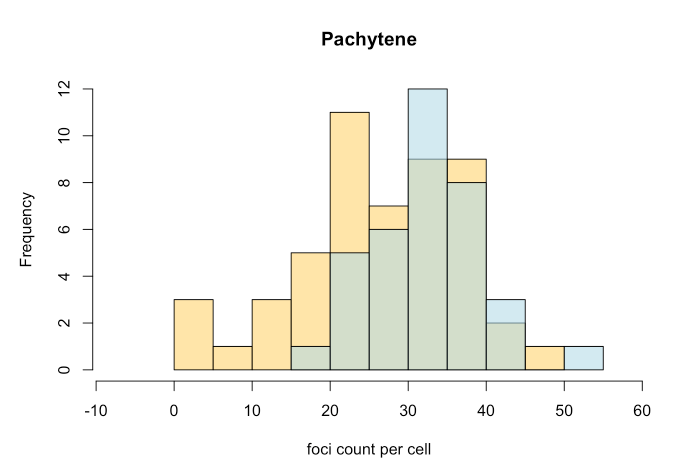 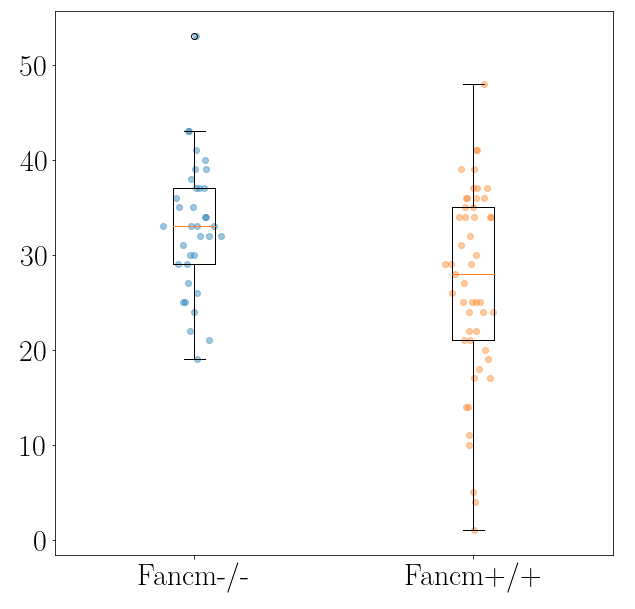 <img src="output/measure_distances_histogram.png" width="700" height="500"> <img src="output/measure_distances_boxplot.png" width="600" height="500"> ## Project organisation and management Please issue bug reports through GitLab. ## Acknowledgements This project is a [workflowr][] project, where we make use of a [project template][] created by Davis McCarthy. [workflowr]: https://github.com/jdblischak/workflowr [project template]: https://gitlab.svi.edu.au/biocellgen-public/aaaa_2019_project-template